News Story
Cell Membrane Structure and Dynamics Modeled with Atomic Resolution

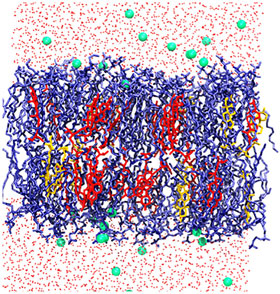
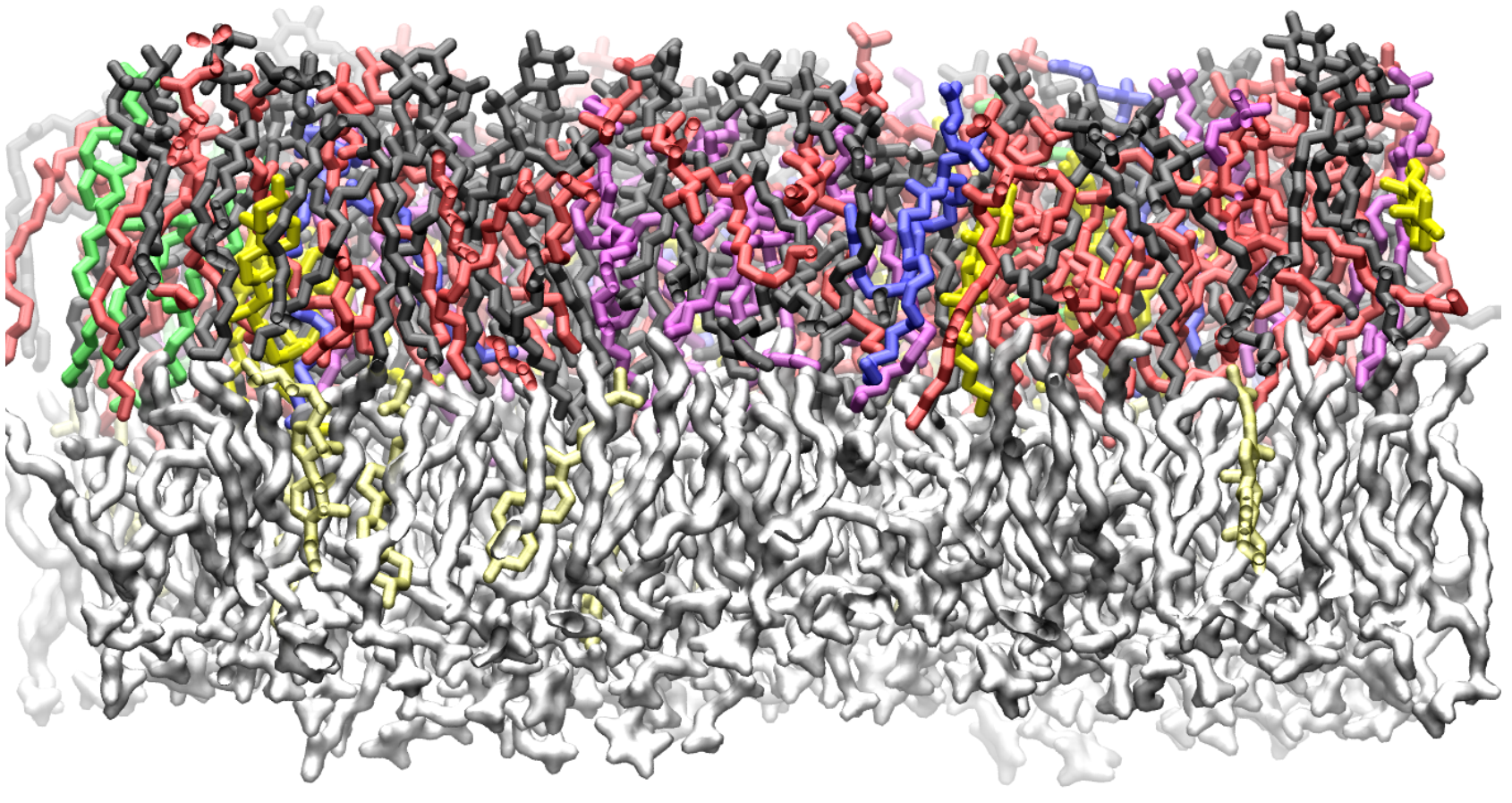
There are thousands of structurally different, naturally occurring lipids that self-assemble into membranes surrounding cells and their internal structures. Lipids can have different polar head groups, along with a variety of kinks and branches in their nonpolar hydrocarbon tails. Now, with developments in molecular modeling, researchers are learning how lipid structural diversity relates to membrane properties and function.
A new perspective from Jeffery Klauda [– a professor of Chemical and Biomolecular Engineering (ChBE) at the University of Maryland (UMD) –] describes how lipid models have grown from describing bilayers containing one lipid type in the early 1990s to current state-of-the art models that describe about 10 different lipids in a bilayer at atomic resolution. The key to including different lipids in a model is an accurate mathematical function, called a force field, which represent the intra- and intermolecular interactions of each lipid. Molecular dynamics simulations using these force fields can predict the structure and dynamics of a model membrane.
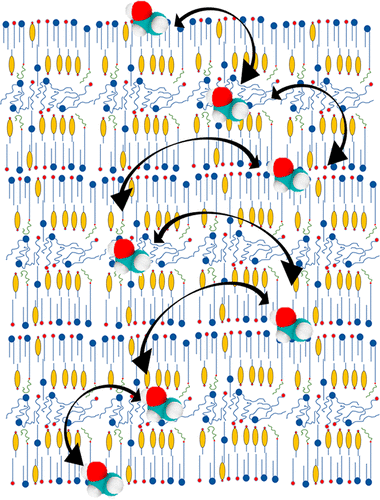
In these models, researchers often randomly combine lipids in a prebuilt bilayer and then let the model equilibrate the membrane structure. Then they use a simulation program to explore how the lipid composition and motion influences membrane structure, physical properties, and interactions with embedded proteins. For example, researchers have modeled a cell membrane from yeast, bacteria, soybeans and the lens in a human eye.
[“The research focuses on the evolving field of modeling realistic cellular membranes,” said Klauda. “The field is at the point to be able to reasonably match the properties of in vivo membranes to study various aspects of these membranes, e.g., how drugs can permeate membranes, how protein function is affected by the varying membrane environment, and how lipid composition influences the mechanical properties of membranes.”]
More coarse grained models allow researchers to simulate larger sections of membrane containing about 100 different lipid types. These models provide insight into how asymmetrical lipid compositions or concentrations affect membrane properties.
The trend for the future of the field, Klauda predicts, is to eventually model an entire cell membrane of a simple organism.
Source: “Perspective: Computational modeling of accurate cellular membranes with molecular resolution,” Jeffery B. Klauda, The Journal of Chemical Physics (2018). The article can be accessed at https://doi.org/10.1063/1.5055007.
*Reprinted with permission from AIP Scilight. Credit: Melissae Fellet, for AIP.
Published January 16, 2019