News Story
UMD Research Offers Insight into Structure of Cellular Membrane
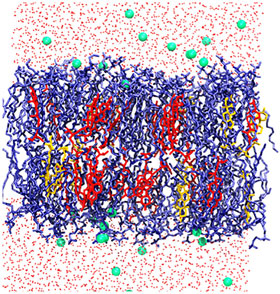
Snapshot of a hypocotyl (stem of a germinating seedling) at the end of the simulation: the glycerol phospholipids are shown in blue; sitosterol and stigmasterol are shown in red and yellow; water is shown in red dots, and green dots are potassium ions.
All living things, regardless of size, be it human, animal or plant, are made up of cells: the so-called ‘building blocks of life.’ Each cell is made up of cytoplasm, the inner material, and the outer plasma membrane, which acts as a protective barrier. The structure of a cell's plasma membrane consists of lipids and proteins, which vary across organisms. This protein/lipid make-up sends signals from cell-to-cell and defines, for example, how flexible it may be, and how permeable; thus, how susceptible a cell may be to viruses, or drug molecules, for example.
Indeed, molecular dynamics computer simulations are now being used to study lipids and proteins as they provide a more comprehensive look at plasma membranes and their abilities. Jeffrey Klauda, an Associate Professor in the UMD Department of Chemical and Biomolecular Engineering (ChBE), and his team have developed a computer simulation of a soybean plasma membrane. To date, soy membranes have not been often studied using computational methods. The hope is that this research will be useful in future projects geared towards the engineering of bio-chemicals and fuels, pharmaceuticals, etc.
In this field, “we have the ability to simulate and probe biologically relevant membranes,” Klauda told the American Institute of Physics. “If you want to understand the structure of a membrane, you really need to include the diversity that exists in biology.”
This research, entitled "Simulations of simple linoleic acid-containing lipid membranes and models for the soybean plasma membranes" (DOI: 10.1063/1.4983655), was published in the Journal of Chemical Physics on June 7, 2017.
Klauda’s research team includes first author Xiaohong Zhuang (who completed her Ph.D. in Dec. ’16) and Anna Ou. Ou was a student at Montgomery Blair High School while this research was being conducted.
Abstract:
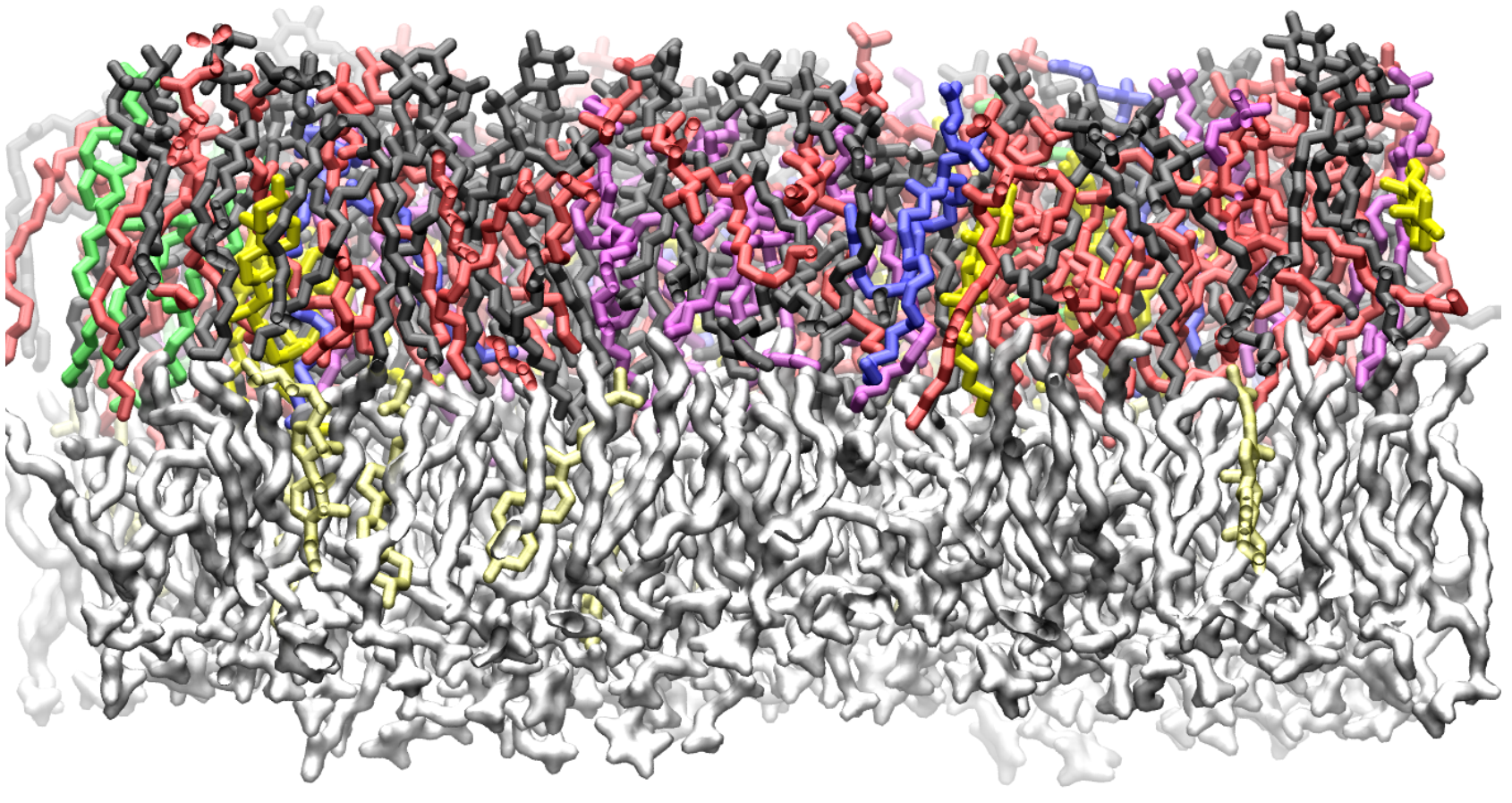
The all-atom CHARMM36 lipid force field (C36FF) has been tested with saturated, monounsaturated, and polyunsaturated lipids; however, it has not been validated against the 18:2 linoleoyl lipids with an unsaturated sn-1 chain. The linoleoyl lipids are common in plants and the main component of the soybean membrane. The lipid composition of soybean plasma membranes has been thoroughly characterized with experimental studies. However, there is comparatively less work done with computational modeling. Our molecular dynamics (MD) simulation results show that the pure linoleoyl lipids, 1-stearoyl-2-linoleoyl-sn-glycero-3-phosphocholine (18:0/18:2) and 1,2-dilinoleoyl-sn-glycero-3-phosphocholine (di-18:2), agree very well with the experiments, which demonstrates the accuracy of the C36FF for the computational study of soybean membranes. Based on the experimental composition, the soybean hypocotyl and root plasma membrane models are developed with each containing seven or eight types of linoleoyl phospholipids and two types of sterols (sitosterol and stigmasterol). MD simulations are performed to characterize soybean membranes, and the hydrogen bonds and clustering results demonstrate that the lipids prefer to interact with the lipids of the same/similar tail unsaturation. All the results suggest that these two soybean membrane models can be used as a basis for further research in soybean and higher plant membranes involving membrane-associated proteins.
Published June 19, 2017